Team: Yongsoo Kim, Perry Moerland, Jos Poell, Betty Tijms
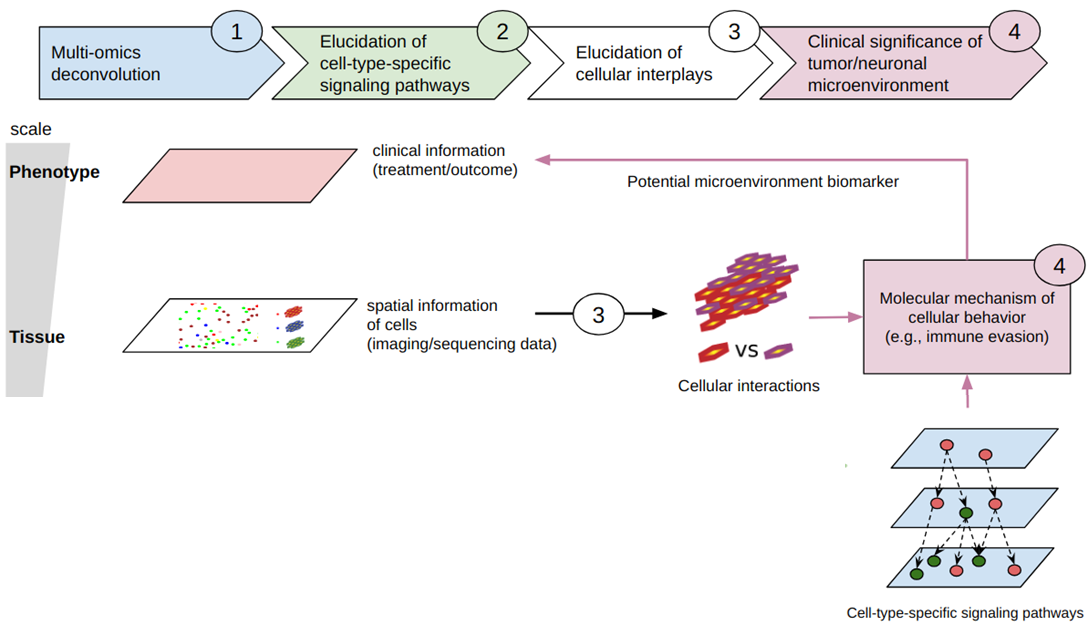
Achieving precise patient care requires an accurate understanding of the mechanisms underlying disease development and drug resistance, but these processes are often heavily dependent on complex interplays of cells within their microenvironment. Cellular heterogeneity can be attributed to cell type composition
in the microenvironment, activation states, and their function in terms of interactions with other cell types (e.g., immune cells and tumour cells; immune cells and
microglia). While advanced single-cell molecular and imaging data can address some aspects of this complexity, their availability is limited due to high cost and cannot be expanded to large series which is necessary for biomarker discovery. Furthermore, these techniques individually cannot cover all of the modalities (e.g., proteome) as efficiently as others (e.g., transcriptome), which makes it challenging to provide a complete picture. Our goal is to overcome these limitations by establishing a comprehensive computational framework that integrates diverse omics data types,
including (single-cell) RNA/DNA sequencing and spatially resolved molecular data, available from both public sources and ongoing projects of Adore researchers. This framework will allow us to comprehensively characterise individual cell types through multi-omic profiles, understand their activation states, and explore spatial domains and community formation. We anticipate that this framework will unveil the intricate cellular heterogeneity in cancer and the brain, ultimately contributing to clinically relevant cellular biology.